The ongoing race to find Aphanomyces resistance |
| By Deborah Samac |
|
|
The author is with USDA-ARS Plant Science Research Unit, St. Paul, Minn.  Sorting out alfalfa seedling diseases that are active in wet soils has proven to be a challenge, but progress is being made. Alfalfa is frequently described as a crop that is not tolerant of wet soils. In the early 1900s, researchers described root rots of alfalfa that were attributed to wet soils or a high water table and advised farmers to avoid these fields or to tile drain wet areas for alfalfa production. The agents causing the disease were later identified, most of them belonging to the group of organisms known as oomycetes or “water molds” that flourish in wet soils and infect plants by means of swimming spores called zoospores. A major culprit of alfalfa root rot is Phytophthora medicaginis, which is found in nearly every region of the world where alfalfa is grown. USDA Agricultural Research Service scientists released “Agate” in 1973, which was the first cultivar with resistance to Phytophthora root rot. Commercial alfalfa breeding companies developed additional resistant varieties, and there is currently a very high level of resistance to this disease in all dormancy groups. Once cultivars with resistance to Phytophthora root rot were widely planted, it became apparent that another disease was also causing root rot in wet soil conditions. Symptoms included thin new plantings with stunted seedlings that developed yellow leaves and had light to dark brown soft, rotted roots. Established plants also were affected during extended periods of wet soil conditions and were found to have rotted lateral roots, lack root nodules, and have stunted and yellow foliage. The culprit identified To determine what was causing the disease symptoms, researchers needed to obtain the pathogen in a pure culture. Attempts to isolate the pathogen from pieces of rotted root tissue were unsuccessful due to the large number of other organisms that colonized the dead roots. Another technique was developed using soil sampled from fields with the disease to grow alfalfa seedlings as “bait.” The pathogen was isolated from seedlings in which the disease was just beginning and was grown successfully on laboratory media. Another oomycete, called Aphanomyces euteiches, was found to be the organism causing this root rot. Using the isolated pathogen to artificially inoculate alfalfa seedlings, researchers at the University of Wisconsin identified resistant plants from a number of alfalfa cultivars. Resistant plants from these tests and healthy plants selected under field conditions were intercrossed to create the first alfalfa lines with Aphanomyces root rot resistance called WAPH-1. The first resistant cultivars were commercially available in the 1990s, and field tests showed that the new cultivars provided dramatically improved seedling health, yield, and persistence. While P. medicaginis is relatively specific to alfalfa, A. euteiches causes serious disease in pea, bean, and red clover as well as alfalfa. The pathogen is composed of a complex of genetically diverse subspecific types. One level of complexity is for host preference. For example, most strains isolated from pea will infect alfalfa but cause more serious symptoms on pea. Some strains isolated from alfalfa will infect pea, but others will not. The second level of complexity is based on race within the alfalfa-infecting types.  Pythium and Fusarium strains cause pre- and postemergence damping off and seed rot of alfalfa (right) compared to soil lacking the pathogens (left). Resistance sometimes failed Distinct races of A. euteiches were identified that infect alfalfa after the release of Aphanomyces root rot resistant cultivars. Not long after the widespread use of these cultivars, there were reports of failures in disease resistance. Aphanomyces root rot was occurring even though plants had been bred for high levels of resistance. Using the alfalfa seedling baiting technique, researchers isolated strains of A. euteiches from field soils that had shown failure of resistant cultivars and used them to inoculate alfalfa seedlings. The new isolates did indeed cause root rot disease on cultivars previously thought to be resistant. New programs of selection at the University of Wisconsin were initiated to identify alfalfa plants with resistance to both types of A. euteiches. These efforts resulted in development of an alfalfa line with resistance to both types called WAPH-5. With two kinds of resistant alfalfa lines, races of the pathogen could be identified. Those that caused disease on Agate but not on WAPH-1 were called race 1, while those that caused disease on WAPH-1 but not on WAPH-5 were called race 2. Commercial alfalfa breeding companies began to develop cultivars with resistance to both race 1 and race 2. Tests with individual soil samples found that race 2 occurs in many areas of the United States. However, it was not clear if it was a major or minor component of the pathogen complex. Starting about 10 years ago, intensive surveys were done in Wisconsin, Minnesota, and New York to determine the distribution of race 1 and race 2 strains. Soil samples were obtained from commercial production fields and used in a bioassay with alfalfa lines susceptible to both Phytophthora and Aphanomyces root rot, “Agate” with Phytophthora root rot resistance, WAPH-1, and WAPH-5. In the three states, it appears that race 2 strains are more common than race 1 strains. That is, the WAPH-5 seedlings were healthy, while the WAPH-1 seedlings were killed. However, there were also a number of “anomalous” soils in which a clear result could not be obtained, suggesting the possibility of additional races. More disturbing were some reports of failures of race 2 resistant varieties in some locations. The search continues Researchers in my laboratory have taken a closer look at the pathogens in the anomalous soil samples. We used the baiting technique to isolate pathogens from soils in which a very high level of seed rot and damping off was observed in the Aphanomyces bioassay. In those soils, almost all seedlings of all the alfalfa lines died but did not have the typical Aphanomyces root rot symptoms. We isolated eight species of Pythium, another oomycete known to cause seed rot and damping off of alfalfa, and showed that three of them were highly virulent on alfalfa. We also isolated four species of Fusarium, a fungal pathogen, which caused pre-emergence rot of alfalfa seeds. Since the 1980s, most alfalfa seed has been treated with the fungicide Apron or ApronXL to control Pythium seed rot and damping off. However, the active ingredient in ApronXL did not inhibit many of the Pythium strains we isolated. Taken together, these results indicate that some of the anomalous bioassay results were due to seed rot and damping off caused by Pythium and Fusarium species. From soils in which WAPH-5 had low levels of resistance, we isolated 60 A. euteiches strains and inoculated the individual strains onto seedlings of WAPH-1 and WAPH-5. Each strain could be clearly identified as either race 1 or race 2 in this test. However, this assumes that WAPH-5 has a single gene for resistance to race 2. If instead, WAPH-5 has many genes for resistance to Aphanomyces root rot, we would not be able to distinguish additional races. Researchers at the University of Wisconsin have evidence for multiple resistance genes in what we have called race 2 resistant lines such as WAPH-5 and evidence for multiple races of A. euteiches. Obtaining lines of alfalfa with single resistance genes is complicated by the outcrossing tetraploid nature of alfalfa. Until we can identify resistance genes in alfalfa at the DNA sequence level, the number of races of A. euteiches will be difficult to determine. On a practical basis, it does not appear that these additional races are causing widespread breakdown of resistance to Aphanomyces root rot in current commercial cultivars. However, to maintain protection against this disease, a wide range of isolates should be used in breeding programs when developing new cultivars. Growers are advised to use the most advanced disease resistant cultivars available. Protecting the alfalfa root from organisms causing decay is of the utmost importance to ensure high yields, stand persistence, and winter survival. 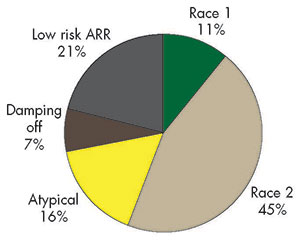 This article appeared in the August/September 2016 issue of Hay & Forage Grower on page 6 and 7. Not a subscriber? Click to get the print magazine. |
